lr test r package|r likelihood test : solutions Conduct the likelihood-ratio test for two nested extreme value distribution models. WEB3. Tinychat. O único ponto semelhante que este site tem com o Omegle é que você pode se comunicar com pessoas aleatórias. No geral, o Tinychat tem um foco maior em comunidade, permitindo que .
{plog:ftitle_list}
Shrimp • Crab • Cucumber • Green Onion •. Carrot • Edamame • Avocado • Soy Sauce •. Tempura Flakes • Spicy Mayo Drizzle.
r likelihood test
Description. lrtest is a generic function for carrying out likelihood ratio tests. The default method can be employed for comparing nested (generalized) linear models (see details below). .Conduct the likelihood-ratio test for two nested extreme value distribution models.
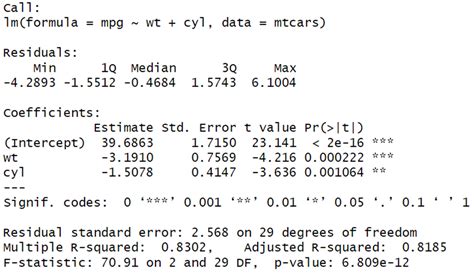
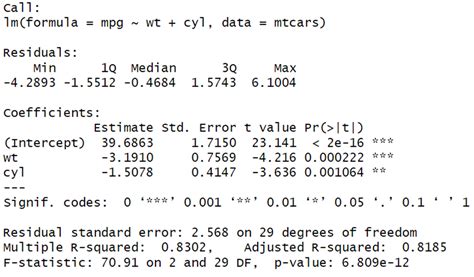
We will use the lrtest () function from the lmtest package to perform a likelihood ratio test on these two models: library(lmtest) . #fit full model . model_full <- lm(mpg ~ disp + .
the heel drop test
Conduct the likelihood-ratio test for two nested extreme value distribution models.This function performs likelihood-ratio tests for the likelihood of the record indicators I t to study the hypothesis of the classical record model (i.e., of IID continuous RVs).Likelihood Ratio Test of the Proportional Odds Assumption Description. Provides the means of testing the parallel regression assumption in the ordinal regression models. Also available is .An alternative is the lmtest package, which has an lrtest() function which accepts a single model. Here is the example from ?lrtest in the lmtest package, which is for an LM but there are .
Description. Conduct the likelihood-ratio test for two nested extreme value distribution models. Usage. lr.test(x, y, alpha = 0.05, df = 1, .) Arguments. Details. Description. Conduct the likelihood-ratio test for two nested extreme value distribution models. Usage. lr.test(x, y, alpha = 0.05, df = 1, .) Arguments. Details.
Likelihood ratio test checks the difference between -2*logLikelihood of the two models against the change in degrees of freedom using a chi-squared test. It is best applied .Description. Test the null hypothesis that the two models fit the data equally well. Usage. LRTest(full, reduced, df = 1, boundaryCorrection = FALSE) Value. a list: lambda. a numeric log .
The function performs a likelihood ration test for two nested fitted model. Rdocumentation. powered by. Learn R Programming. gamlss (version 5.4-12) Description. Usage Value, Arguments. Author. Details . (y~x1+x2+x3+x4, data=usair) LR.test(m0,m1) Run the code above in your browser using .Test the null hypothesis that the two models fit the data equally well. Rdocumentation. powered by. Learn R Programming. nadiv (version 2.18.0) Description. Usage Value: Arguments. Author. Details. References. See Also. Examples Run this code # No boundary correction (noBC <- LRTest(full = - 2254.148, reduced = - 2258.210, df = 1 . rdrr.io Find an R package R language docs Run R in your browser. lmtest Testing Linear Regression Models. . Likelihood Ratio Test of Nested Models Description. . data = usdl) fm2 <- lm(con ~ gnp + con1 + gnp1, data = usdl) ## various equivalent specifications of the LR test lrtest(fm2, fm1) lrtest(fm2, 2) lrtest(fm2, "con1") lrtest(fm2 .Compute likelihood ratio test to compare two fitted models, one nested within the other. Rdocumentation. powered by. Learn R Programming. secr (version 4.6.6) Description. Usage Value . LR.test (secrdemo.0, secrdemo.b) Run the code above in your browser using .
Compute score tests comparing a fitted model and a more general alternative model. To compare nested models, you can use different criteria including p-value from LRT or ANOVA, Adjusted-R 2, AIC, BIC and so on.LRT and ANOVA would yield the same outcome in terms of detecting a difference.
LR-test p-value (invisibly) . R Package Documentation. rdrr.io home R language documentation Run R code online. Browse R Packages. CRAN packages Bioconductor packages R-Forge packages GitHub packages. We want your feedback! Note that we can't provide technical support on individual packages. You should contact the package authors for .
By Wilk’s Theorem we define the Likelihood-Ratio Test Statistic as: λ_LR=−2[log(ML_null)−log(ML_alternative)] Why is it true that the Likelihood-Ratio Test Statistic is chi-square distributed? First recall that the chi-square distribution is the sum of the squares of k independent standard normal random variables. Below is a graph of the .
Computes the likelihood ratio test for the coefficients of a generalized linear model. Rdocumentation. powered by. Learn R Programming. mdscore (version 0.1-3) . ("inverse")) lr.test(fit0,fitf) Run the code above in your browser using .Likelihood Ratio Test of Nested Models Description. lrtest is a generic function for carrying out likelihood ratio tests. The default method can be employed for comparing nested (generalized) linear models (see details below). We can also plot these values to visualize and get a better understanding using the "ggplot2" package in the R programming Language. R . In R, the T-test can be extended to handle multiple groups by using approaches like pairwise comparisons or ANOVA (Analysis of Variance). This artic. 4 min read. How to Perform Paired t-Test for Multiple .Hosmer-Lemeshow Goodness of Fit (GOF) Test.
This function performs likelihood-ratio tests for the likelihood of the record indicators \(I_t\) to study the hypothesis of the classical record model (i.e., of IID continuous RVs). rdrr.io Find an R package R language docs Run R in your browser. mdscore Improved Score Tests for Generalized Linear Models . Functions. 12. Source code. 2. Man pages. 5. lr.test: Likelihood ratio test for generalized linear models; mdscore: Modified score test for generalized linear models; strength: Impact Strength an Insulating Material . Details. The null hypothesis of the likelihood-ratio tests is that in every vector (columns of the matrix X), the probability of record at time t is 1 / t as in the classical record model, and the alternative depends on the alternative and probabilities arguments. The probability at time t is any value, but equal in the M series if probabilities = "equal" or different in the M series if .
Likelihood ratio test checks the difference between -2*logLikelihood of the two models against the change in degrees of freedom using a chi-squared test. It is best applied to a model from 'glm' to test the effect of a factor with more than two levels. The records used in the dataset for both models MUST be the same. rdrr.io Find an R package R language docs Run R in your browser. VAR.etp VAR Modelling: Estimation, Testing, and Prediction. Package index. . LR test statistic. pval: p-value of the LR test. Boot.pval: p-value of the test based on bootstrapping. Note. See Chapter 4 of Lutkepohl (2005) Author(s)
Note. Both fit1 and fit2 must have the same family and link function.. Author(s) Damiao N. da Silva [email protected]. Antonio Hermes M. da Silva-Junior [email protected]. References. McCullagh P, Nelder J (1989). Generalized Linear Models.Chapman & Hall/CRC, London.null: The null hypothesis (simpler) fitted model alternative: The alternative hypothesis (more complex) fitted model print: whether to print or save the result
lrtest is a generic function for carrying out likelihood ratio tests. The default method can be employed for comparing nested VGLMs (see details below).The lmerTest package provides p-values in type I, II or III anova and summary tables for linear mixed models ( lmer model fits cf. lme4 ) via Satterthwaite's degrees of freedom method; a Kenward-Roger method is also available via the pbkrtest package. . ## Inspect the contrast matrix for the Type III test of Product: show_tests(aov, fractions .
Details. The null hypothesis of the likelihood-ratio tests is that in every vector (columns of the matrix X), the probability of record at time t is 1 / t as in the classical record model, and the alternative depends on the alternative and probabilities arguments. The probability at time t is any value, but equal in the M series if probabilities = "equal" or different in the M series if .
Computes a Wald \(\chi^2\) test for 1 or more coefficients, given their variance-covariance matrix.Details. lrtest is intended to be a generic function for comparisons of models via asymptotic likelihood ratio tests. The default method consecutively compares the fitted model object object with the models passed in ..Instead of passing the fitted model objects in ., several other specifications are possible.The updating mechanism is the same as for waldtest() in lmtest: .u-test SI LR - Integrating Package Monitoring Indicators - 5000 Pack Item 81121 5000 integrators per bag: 8.50. per bag. Add to Cart. Never purchased me before? Try me for just Instead of a Ftest comparing the SSR for the univariate case, a Likelihood Ratio (LR) test comparing the covariance matrix of each model is computed. LR_{ij}=T( ln(\det \hat \Sigma_{i}) -ln(\det \hat \Sigma_{j})) where \hat \Sigma_{i} is the estimated covariance matrix of the model with i regimes (and so i-1 thresholds). Three test are available..00 * u-test SI LR - Integrating Package Monitoring Indicators - 10 000 Pack Item 81127 .The LR.Sarlm() function provides a likelihood ratio test for objects for which a logLik() function exists for their class, or for objects of class logLik . LR1.Sarlm() and Wald1.Sarlm() are used internally in summary.Sarlm() , but may be accessed directly; they report the values respectively of LR and Wald tests for the absence of spatial .
the human egg drop test
WEBAcompanhe os Resultados do Jogo do Bicho FEDERAL de 19 horas válido para todo o país, os sorteios são realizados as Quartas e Sábados as 19:00 de Brasília. Aproveite e .
lr test r package|r likelihood test